In vivo, in vitro, in silico – rule of three for Systems Biology
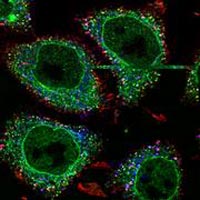
Prof. Marino Zerial searches after visualisation of cell organells, to make them accesible for modeling with the computer Photo: M. Zerial <br>"Reprinted from Cell, Vol.116 , Authors: M. Zerial, Miaczynska, Title of article: APPL Proteins Link Rab5, Pages 445 - 456, Copyright 06.02.2004 with permission from Elsevier
Prof. Jens Reich, Biomathematician and member of the German National Ethics Council, is one of the scientists within the Systems Biology competence network HepatoSys* who is on the scent of a major mystery of life. His main concern is the question how biological systems with a vast number of subprocesses can function. In order to understand the linkages, identifying agents such as proteins and encymes does not suffice. Of equal importance is the exploration of their interdependency.
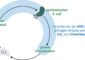
The close collaboration between scientists working in experimental medicine and biology with theorists doing computer modeling in mathematics, physics and engineering sciences has already stood the test. Two years after the launch of the BMBFs funding focus Systems Biology, the networkers of HepatoSys have placed an initial milestone on the way to a virtual liver cell. The aim to reproduce physiological processes not only in vivo (in living organisms) and in vitro (in the test tube), but also in silico** (in the computer), has come closer.
First results
The scientists have succeeded in building a mathematical model of the socalled JAK-STAT signal cascade which contributes to the regulation of both the development and the growth control in mammalian cells. With the help of this model they can predict how fast single steps in signal transduction will take place. Surprisingly, the signal pathway is not linear but occurs in a cyclical manner which is the base for a finely tuned signal transduction.
„The analysis in hepatocytes shows that this principle is universally valid for the JAK-STAT signal pathway. As opposed to conventional approaches, our in silico analyses illustrate that stages of the molecule transport into and out of the nucleus are best suited for an inhibition of signal transduction“ explains Dr. Ursula Klingmüller of the HepatoSys network Regeneration. Predictions developed with the help of computers could be validated experimentally. This way, by using the Systems Biology approach, valuable information regarding biological questions can be gathered. They can be used beyond liver cell research, e.g. in the development of cancer medication.
The Vision
Systems Biology is effective yet still at the beginning. Clinical application is a long way ahead says Prof. Timmer, scientific speaker of HepatoSys. „It is our longterm goal to advance medicine from a qualitative, descriptive research discipline to a quantitatively predictive research discipline.“
* HepatoSys: a neologism combining Hepatocyte – liver cell – and Systems Biology
** in silico: named after the chemical element silicon which is the base for the production of computer chips. In Systems Biology, the term means the evaluation and graphical display of experimentally gained computer data.
Contact:
Dr. Ute Heisner
HepatoSys –
Network Systems Biology
Project Management
Institute for Physics
University Freiburg
Phone: +49 – (0)761 – 203 5803
Fax: +49 – (0)761 – 203 5967
Email: ute.heisner@fdm.uni-freiburg.de
Media Contact
Weitere Informationen:
http://www.systembiologie.deAlle Nachrichten aus der Kategorie: Biowissenschaften Chemie
Der innovations-report bietet im Bereich der "Life Sciences" Berichte und Artikel über Anwendungen und wissenschaftliche Erkenntnisse der modernen Biologie, der Chemie und der Humanmedizin.
Unter anderem finden Sie Wissenswertes aus den Teilbereichen: Bakteriologie, Biochemie, Bionik, Bioinformatik, Biophysik, Biotechnologie, Genetik, Geobotanik, Humanbiologie, Meeresbiologie, Mikrobiologie, Molekularbiologie, Zellbiologie, Zoologie, Bioanorganische Chemie, Mikrochemie und Umweltchemie.
Neueste Beiträge

Bakterien für klimaneutrale Chemikalien der Zukunft
Forschende an der ETH Zürich haben Bakterien im Labor so herangezüchtet, dass sie Methanol effizient verwerten können. Jetzt lässt sich der Stoffwechsel dieser Bakterien anzapfen, um wertvolle Produkte herzustellen, die…

Batterien: Heute die Materialien von morgen modellieren
Welche Faktoren bestimmen, wie schnell sich eine Batterie laden lässt? Dieser und weiteren Fragen gehen Forschende am Karlsruher Institut für Technologie (KIT) mit computergestützten Simulationen nach. Mikrostrukturmodelle tragen dazu bei,…

Porosität von Sedimentgestein mit Neutronen untersucht
Forschung am FRM II zu geologischen Lagerstätten. Dauerhafte unterirdische Lagerung von CO2 Poren so klein wie Bakterien Porenmessung mit Neutronen auf den Nanometer genau Ob Sedimentgesteine fossile Kohlenwasserstoffe speichern können…